Usage
Input
When submitting a job to the waste waters pipeline, you will be presented with the following screen:
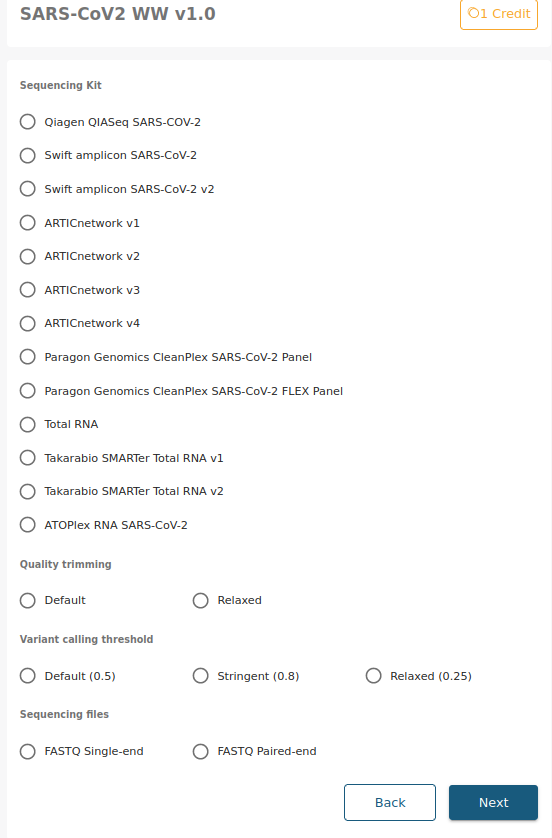
Options
Sequencing kit
Here you can select the sequencing kit used to obtain the data. We currently have the following sequencing kits available, but if the kit your are looking for is not in the list, do not hesitate to contact us and we will add it as soon as possible.
Qiagen QIASeq SARS-COV-2
Swift amplicon SARS-CoV-2
Swift amplicon SARS-CoV-2 v2
ARTICnetwork v1
ARTICnetwork v2
ARTICnetwork v3:
ARTICnetwork v4
Paragon Genomics CleanPlex SARS-CoV-2 Panel
Paragon Genomics CleanPlex SARS-CoV-2 FLEX Panel
Total RNA
Takarabio SMARTer Total RNA v1
Takarabio SMARTer Total RNA v2
ATOPlex RNA SARS-CoV-2
Quality trimming
Here you can select the stringency of the quality trimming step. The Default parameter will trim the reads where the mean quality drops below 30, and the Relaxed parameter will trim the reads where the mean quality drops below 20.
Variant calling threshold
Here you can select the stringency of the variant calling step. The Default (0.5) parameter will call a variant when the allelic frequency of the variant is 0.5 or greater, the Stringent (0.8) parameter will call a variant when the allelic frequency of the variant is 0.8 or greater, and the Relaxed (0.25) parameter will call a variant when the allelic frequency of the variant is 0.25 or greater.
Sequencing files
Select whether your data is single-end or paired-end.
Accepted sample names
Stratus web server uses the last characters on the file name to determine which mate is which in paired-end data. This is why the accepted file terminations are the following:
_1.fastq.gz and _2.fastq.gz
_1.fq.gz and _2.fq.gz
For single-end samples the accepted terminations are the same but without the mate information.
Launching
When clicking Next in the option screen, the file uploading screen will show up. Once the FASTQ files have been selected (either by drag-and-drop or selection via file explored), you can change the name of the samples.
Once all the options have been selected, by clicking next the sample upload will begin and the analysis pipeline will launch!
An email will be received once the analysis are finished and the results will be available on the web server.